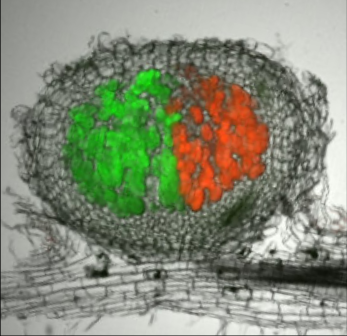
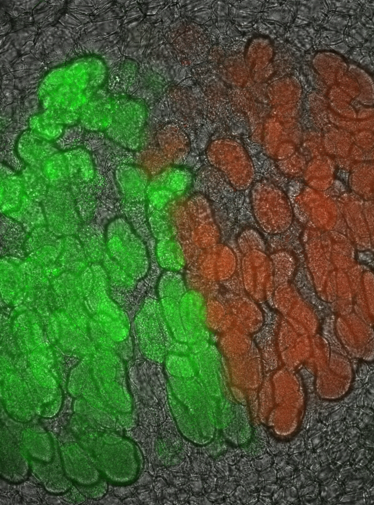
Priority Programme SPP 2125: Deconstruction and Reconstruction of the Plant Microbiota, DECRyPT
Summary
In nature, the roots and leaves of plants engage in intimate associations with an enormous diversity of microbes including bacteria, fungi, algae, oomycetes and protists. Collectively, the microbial assemblage of a plant is called the “plant microbiota” and the sum of the plant-inhabiting microbial genomes the “plant microbiome”. Previous studies have demonstrated not only adverse, but also beneficial functions of individual members of these microbial assemblages for plant hosts, including nutrient mobilisation and uptake, protection against plant pathogens, or abiotic stress tolerance. However, the current shortage of fundamental knowledge on principles underlying community establishment and functions conferred by microbe-microbe and/or microbe-host interactions in these assemblages makes it difficult to predict whether individual members of the microbiota retain their beneficial or detrimental activities in a microbial community context.
The central scientific objectives of this Priority Programme are to obtain a deep and more predictive understanding of plant-microbiota associations and to develop pioneering reductionist approaches towards a molecular understanding of plant microbiota functions. In this Priority Programme we elucidate genetic factors underlying plant microbiota establishment, test presumed community adaptation in ecological contexts and define community-associated emergent properties. Computational and genomic tools will guide hypothesis testing and the design of microbiota reconstitution experiments in controlled environments.
This Priority Programme aims at a pragmatic understanding of the plant microbiota by application of systematic reductionist approaches, including the deconstruction and reconstruction of microbial assemblages. The deconstruction phase involves establishment of model microbial culture collections from plants grown in contrasting natural environments and microbial whole-genome sequencing of pure strains. The reconstruction phase includes microbiota reconstitution experiments using gnotobiotic plant systems to test the impact of different microbes and defined environments on plant fitness parameters such as disease resistance, nutrient acquisition, resource allocation and abiotic stress tolerance under laboratory conditions, to help us understand their roles in nature. One challenge is to determine how the manipulation of specific host and microbial pathways affects microbiota composition, their protective effects and consequently plant health. The ultimate goal is to validate inferences from the reductionist approaches in natural or agricultural ecosystems.
To maximise the potential for synergies and cross-referencing of data, three model plants will be employed in this Priority Programme, the dicotyledonous model Arabidopsis thaliana (and related sister species), the legume symbiosis model Lotus japonicus (and related Lotus corniculatus) and the cereal crop model Hordeum vulgare. Programme members are encouraged to use a standardised natural substrate and characterised microbiota culture collections. We particularly encourage projects, which move from exploratory to mechanistic studies over the course of this Priority Programme. For accurate and high-resolution analysis of microbiome data, the programme has established a central platform providing members with computational and genomic tools and will establish mutant libraries. It also provides access to comprehensive microbe culture collections. The Priority Programme will not cover binary interactions between host plants and a single microbe, the virome of plants, or studies focussing on soil biophysics or soil geochemistry, as this would dilute the focus of the programme on understanding the molecular basis and ecological relevance of plant-associated microbial assemblages.